Similarity Based Functions
In this section, the similarity functions are reviewed with some examples for similarity module. The purpose of the similarity module is to calculate different types of similarity between two or more 3D NifTi images. The types of similarity estimates include the Dice Coefficient, Jaccard Coefficient, the Tetrachoric or Spearman Rank correlation coefficients.
The formulas for each are as follows:
image_similarity
The NifTi images that are used with this function can be from SPM, FSL, AFNI or Nilearn preprocessed outputs. The two requirements the user has to confirm are:
Image Shape: The two images being compared have to be of the same shape. If the images are of different length the the comparison of the volumes will be wrong. The package will throw an error if the images are of the wrong shape.
Normal/Coregistration: The images should be in standard space for appropriate comparison. While the above error should arise, it will not check whether the images are in standard space.
You can check the shape of the data using the following. However, standard space has to be confirmed via your preprocessing pipeline. .. code-block:: python
from nilearn import image # checking image shape img1 = image.load_img(“/path/dir/path_to_img.nii.gz”) img1.shape
If your data does meet the requirements, you can easily use image_similarity(). The requirements for the function are:
imgfile1: this is the string for the path to the first image (e.g., /path/dir/path_to_img1.nii.gz)
imgfile2 this is the string for the path to the first image (e.g., /path/dir/path_to_img2.nii.gz)
mask: The mask is optional, but it is the path to the mask (e.g., /path/dir/path_to_img_mask.nii.gz)
thresh: The threshold is optional but highly recommended for Dice/Jaccard/Tetrachoric, the similarity between unthresholded binary images will usually be one (unless they were thresholded before). Base the threshold on your input image type. For example a beta.nii.gz/cope.nii.gz file would be thresholded using a different interger than zstat.nii.gz
similarity_type: This the similarity calculation you want returned. The options are: ‘dice’, ‘jaccard’, ‘tetrachoric’ or ‘spearman’
Let’s say I want to fetch some data off of neurovault and calculate the similarity between two images. For this example I will use the HCP task group activation maps. Instead of manually downloading and adding paths, you can use nilearn to fetch these maps.
from nilearn.datasets import fetch_neurovault_ids
# Fetch hand and foot left motor map IDs
L_foot_map = fetch_neurovault_ids(image_ids=[3156])
L_hand_map = fetch_neurovault_ids(image_ids=[3158])
You can look at the images to see the activation maps for each image:
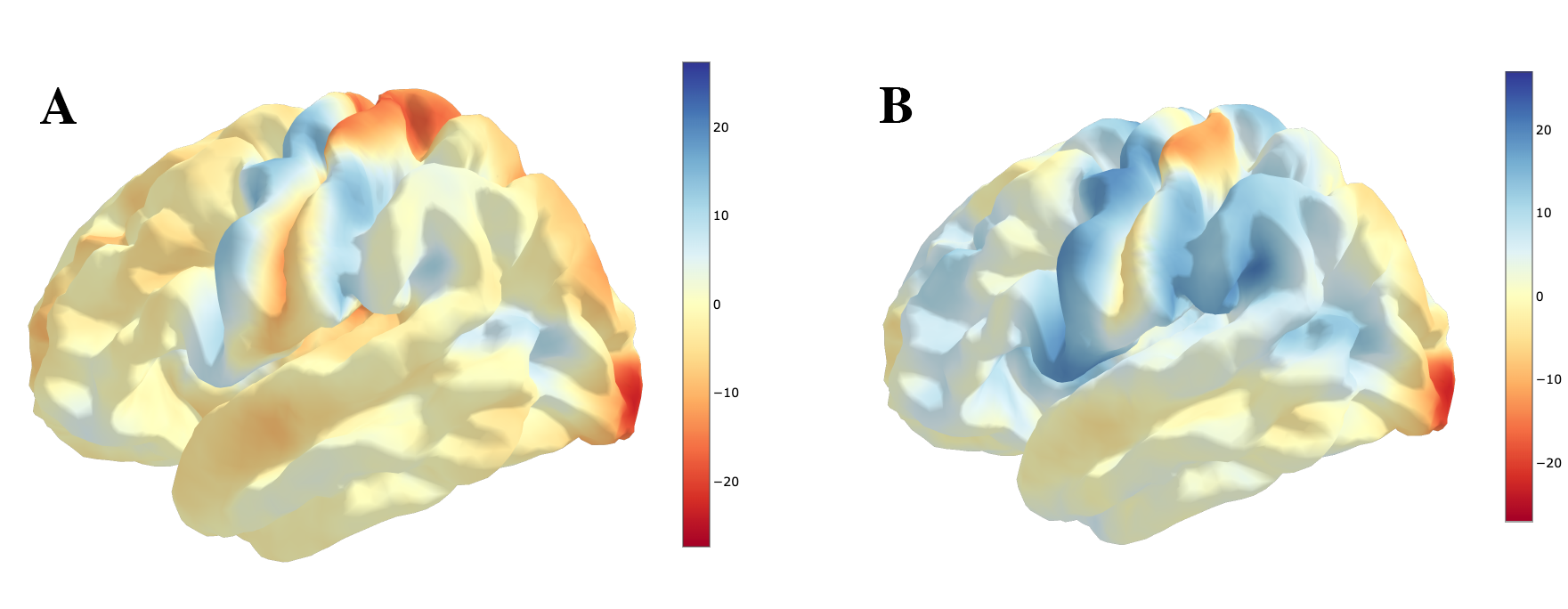
Now that the data are have in the environment, load the similarity package from pyrelimri and calculate the jaccard similarity coefficient and tetrachoric correlation between the two images.
from pyrelimri import similarity
# calculate jaccard coefficient
similarity.image_similarity(imgfile1=L_hand_map.images[0], imgfile2=L_foot_map.images[0], thresh = 1.5, similarity_type = 'jaccard')
similarity.image_similarity(imgfile1=L_hand_map.images[0], imgfile2=L_foot_map.images[0], thresh = 1.5, similarity_type = 'tetrachoric')
The Jaccard coefficient is 0.18 and the Tetrachoric similarity is .776.
Try changing the threshold to 1.0. What would happen? In this instance the correlation will decrease and the Jaccard Coefficient will increase. Why? This is, in part, explained by the decreased overlapping zeros between the binary images and the increased number of overlapping voxels in the Jaccard calculation.
pairwise_similarity
The pairwise_similarity() function is a wrapper for the image_similarity() function within the similarity module. It accepts similar values, except this time instead of imgfile1 and imgfile2 the function takes in a list of paths to NifTi images. The inputs to the pairwise_similarity() function are:
nii_filelist: A list of Nifti files, (e.g., [“/path/dir/path_to_img1.nii.gz”, “/path/dir/path_to_img2.nii.gz”, “/path/dir/path_to_img3.nii.gz”)
mask: The mask is optional but it is the path to a Nifti brain mask (e.g., /path/dir/path_to_img_mask.nii.gz)
thresh: The threshold is optional but highly recommended for binary estimates. The similarity between unthresholded binary images is usually one (unless they were thresholded before). Base the threshold on your input image type. For example a beta.nii.gz/cope.nii.gz file would be thresholded using a different interger than zstat.nii.gz
similarity_type: This the similarity estimate between images that you want returned. The options are: ‘dice’, ‘jaccard’, ‘tetrachoric’ or ‘spearman’
Using the HCP example from above, add two more images into the mix.
from nilearn.datasets import fetch_neurovault_ids
# Fetch hand and foot left motor map IDs
L_foot_map = fetch_neurovault_ids(image_ids=[3156])
L_hand_map = fetch_neurovault_ids(image_ids=[3158])
R_foot_map = fetch_neurovault_ids(image_ids=[3160])
R_hand_map = fetch_neurovault_ids(image_ids=[3162])
I wont plot these images, but for reference there are now four image paths: L_hand_map.images[0], L_foot_map.images[0], R_hand_map.images[0], R_foot_map.images[0]. Now I can try to run the pairwise_similarity() function:
# If you hadn't, import the package
from pyrelimri import similarity
similarity.pairwise_similarity(nii_filelist=[L_foot_map.images[0],L_hand_map.images[0],
R_foot_map.images[0],R_hand_map.images[0]],thresh=1.5, similarity_type='jaccard')
As noted previously, the permutations are across the image combinations. This will return a pandas Dataframe. Such as:
similar_coef |
image_labels |
|
|---|---|---|
0 |
0.18380588591461908 |
image_3156.nii.gz ~ image_3158.nii.gz |
1 |
0.681449273874364 |
image_3156.nii.gz ~ image_3160.nii.gz |
2 |
0.3912509226509201 |
image_3156.nii.gz ~ image_3162.nii.gz |
3 |
0.18500433729643165 |
image_3158.nii.gz ~ image_3160.nii.gz |
4 |
0.2340488091737724 |
image_3158.nii.gz ~ image_3162.nii.gz |
5 |
0.41910546659304254 |
image_3160.nii.gz ~ image_3162.nii.gz |
FAQ
Can I use these function on output from FSL, AFNI or SPM?
Yes, you can use these functions on any NifTi data that are of the same shape and in the same space. You just need the paths to the locations of the .nii or .nii.gz files for the contrast beta, t-stat or z-stat maps.
Are there restrictions on which data I should or shouldn’t calculate similarity between?
It all depends on the question. You can calculate similarity between group level maps or individual maps. There are two things to keep in mind: Ensure the data is in the form that is expected and be cautious about the thresholding that is used because a threshold of 2.3 on a t-stat.nii.gz may not be as restrictive on the group maps as it is on the the individual maps. Note, the spearman estimate is intended to be on raw values and not thresholded values. In this case, thresh should remain as default ‘None’.