pyrelimri
The pyrelimri package contains multiple modules for calculating image reliability measures.
brain_icc
From pyrelimri the brain_icc module contains the following function to calculate voxelwise and atlas based (region of interest) intraclass correlation estimates on provided 3D volumes.
brain_icc.voxelwise_icc(mutlisession_list, mask, icc_type = ‘icc_3’): As shown in Figure 1, calculates the intraclass correlation (e.g., ICC(1), ICC(2,1), or ICC(3,1)) for 3D volumes across 1+ sessions. Returns: five 3D volumes reflecting the estimated ICC, the 95% lowerbound for ICC, 95% upperbound for ICC, mean squared error between subjects & mean squared error within subjects values.
Inputs:
REQUIRED: list of lists to >1 sessions/runs of 3D Nifti images: string (Subjs across Sessions must be in SAME order); Path to 3D Nifti brain mask: string
OPTIONAL: ICC type, icc = string (default = ‘icc_3’, options include: ‘icc_3’,’icc_2’,’icc_1’). Default set to icc_3
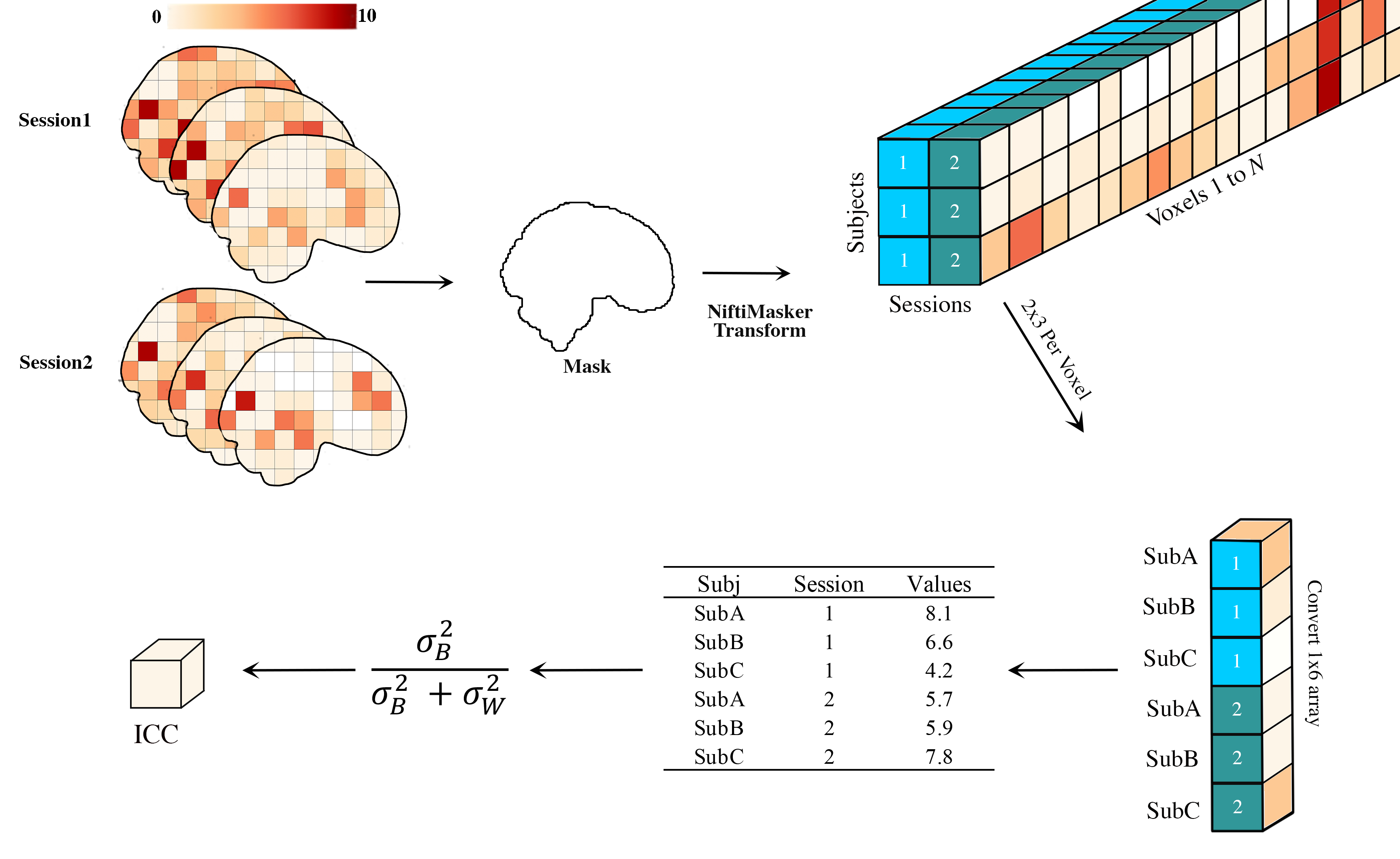
Figure 1. Voxelwise Intraclass Correlation
brain_icc.roi_icc(multisession_list, type_atlas, atlas_dir, icc_type = ‘icc_3’): As show in Figure 2, calculates the intraclass correlation (e.g., ICC(1), ICC(2,1), or ICC(3,1)) based on pre-specified atlas for 3D volumes across 1+ sessions, returning five arrays and five 3D volumes reflecting the ICC estimate, the 95% lowerbound for ICC estimate, 95% upperbound for ICC estimate, mean squared error between subjects, mean squared error within subjects.
Inputs:
REQUIRED: list of lists to >1 sessions/runs of 3D Nifti images: string (Subjects across Sessions must be in SAME order); atlas type ‘aal’, ‘destrieux_2009’, ‘difumo’, ‘harvard_oxford’, ‘juelich’, ‘msdl’, ‘pauli_2017’, ‘shaefer_2018’, ‘talairach’, atlas directory where atlas already exists or should be saved (recommend: ‘/tmp/’), and additional options as required for each atlas at Nilearn datasets. Variable and value, e.g., dimension=64 can be added as instructed in the atlases documentation
OPTIONAL: ICC type, icc = string (default = ‘icc_3’, options include: ‘icc_3’,’icc_2’,’icc_1’). Default set to icc_3.
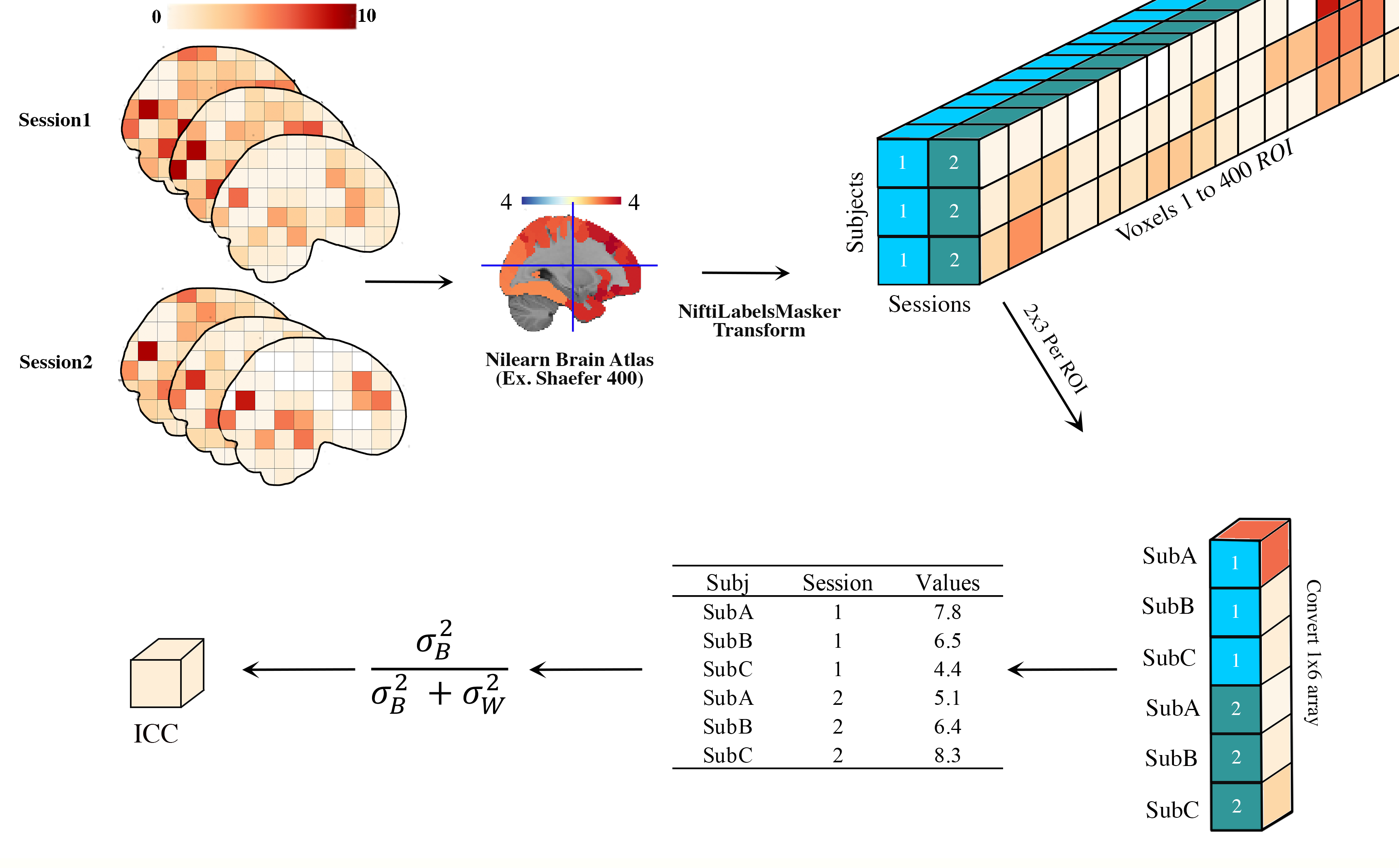
Figure 2. Atlas Based Intraclass Correlation
Note: For deterministic atlases the input data are resampled to the atlas space (e.g., resample_target=’labels’) and probabilistic atlases the atlas is resampled to the data space (e.g., resample_target=’data’). The former is to maintain the boundaries of the ROIs and the latter is to reduce computation time as the probablistic atlas are weighted (this weight results in smoothing of the data). The function will return an array of 1 x ROIs for the ICCs, +/- 95%, MSWS, MSBS and the 3D images that are the 1 x ROIs estimates converted back to the atlas space via inverse_transform.
icc
From pyrelimri the icc module contains the following functions:
icc.sumsq_total(df_long, values): Calculates to total sum of squared error between subjects & sessions.
icc.sumsq_within(df_long, sessions, values, n_subjects): Calculates the sum of squared error within subjects across sessions
icc.sumsq_btwn(df_long, subj, values, n_sessions): Calculates the sum of squared error between subjects
icc.icc_confint(msbs, msws, mserr, msc, n_subjs, n_sess, icc_2 = None, alpha = 0.05, icc_type = ‘icc_3’): Calculates the 95% confidence interval (default) using f-statistic
icc.sumsq_icc(df_long, sub_var, sess_var, value_var, icc_type = ‘icc_3’): Calculates the total, within, between error and returns ICC estimate (e.g., ICC(1), ICC(2,1), or ICC(3,1)) with associated 95% lowerbound and 95% upperbound for ICC, mean between subject variance and mean within-subject variance.
- Inputs:
REQUIRED: Panda long dataframe with: subject variable, sub_var: string; session variable, sess_var: string; the scores, value_var: string;
OPTIONAL: the icc type for sumsq_icc() & icc_confint(), icc_type: string (default = ‘icc_3’, options: ‘icc_3’, ‘icc_2’, ‘icc_1’)
similarity
from pyrelimri the similarity module contains the following functions to calculate the overlap between active/not active voxels for are specified threshold using Jaccard & Dice Coefficient (Fig 3a/3b) or unthreshold voxelwise similarity in activity via spearman rank coefficient (Fig 3d):
similarity.image_similarity(imgfile1, imgfile2, mask = None, thresh = None, similarity_type = ‘dice’): Calculates the similarity between two images. For example, in Figure 3a a measure 1 nifti img1 (thresholded p < 001, blue) and img2 (thresholded p <001, green). The overlapping thresholded voxels are in red. By requesting the Jaccard similarity coefficient (Fig3b), you will get a index of similarity between these two nifti images. Alternatively, you may ask what is the similarity using a binary correlation. Using tetrachoric correlation (Fig3c) we can get the similarity between voxels that are above the p < .001 threshold (==1) and those below (==0) between the two images.
similarity.pairwise_similarity(nii_filelist, mask = None, thresh = None, similarity_type = ‘dice’): Calculates the similarity between two images. Permute across 2+ images to calculate similarity coefficient between all possible image pairs.
- Inputs:
REQUIRED: Path to 3D Nifti, imgfile1: string; Path to 3D Nifti, imgfile2: string; 3D Nifti list, nii_filelist: string
OPTIONAL: Path to a mask: string; threshold on images, thresh: float; similarity type betwee images, similarity_type: string (default = ‘dice’, options include: ‘dice’, ‘jaccard’, ‘tetrachoric’)
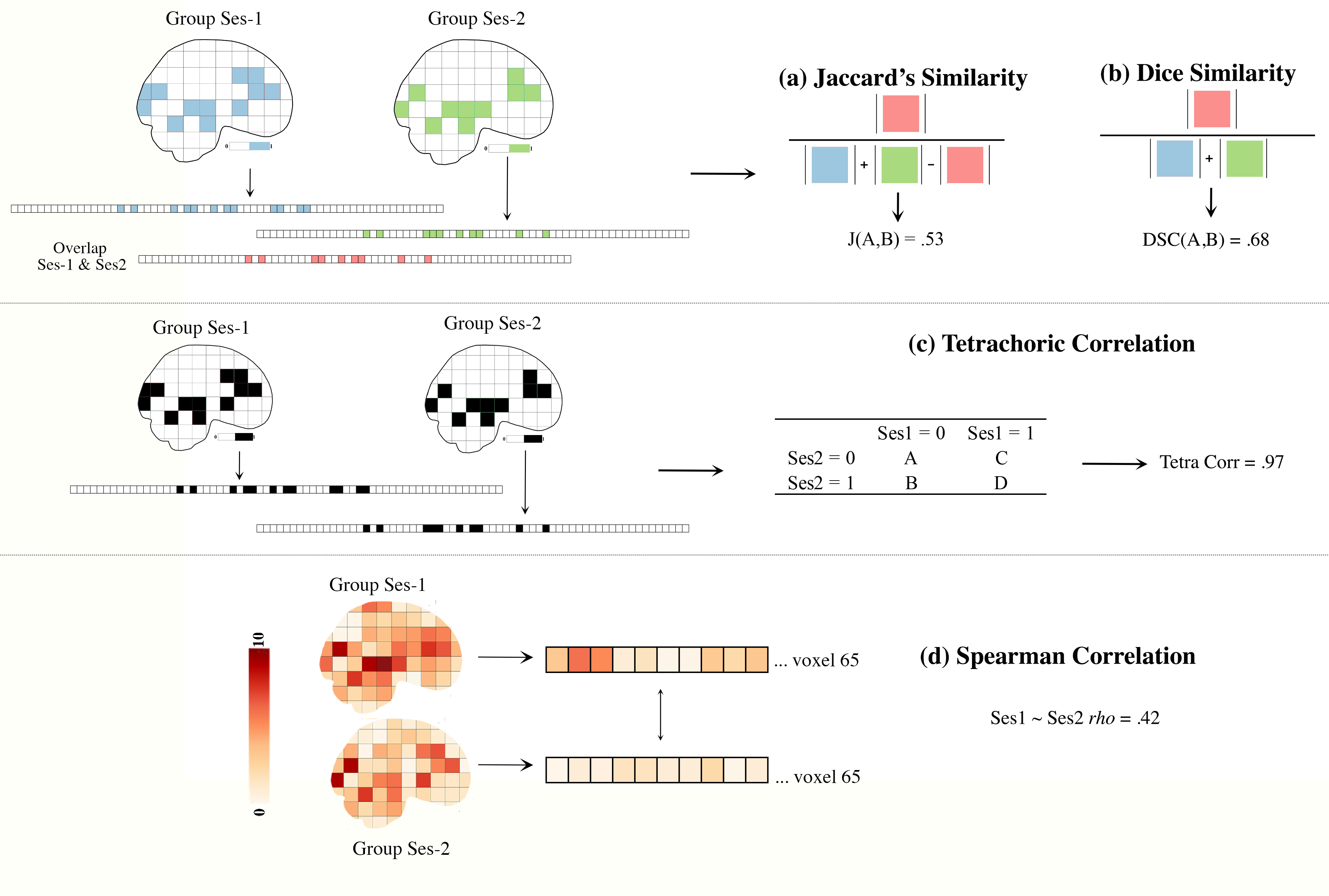
Figure 3. Similarity Between Images
tetrachoric_correlation
From pyrelimri the tetrachoric_correlation module contains the following function to calculate the correlation between two binary images (Fig 3c, 1 = voxels suprathreshold; 0 = voxels subthreshold):
tetrachoric_correlation.tetrachoric_corr(vec1, vec2): Calculates the tetrachoric correlation between two binary vectors.
- Inputs:
REQUIRED: Binary vector1, vec1: NDarray; Binary vector2, vec2: NDarray